Publications
Eeshaan Jain*, Kiril Vasilev*, Alexandre Misrahi*, Phil F Cheng, Petros Liakopoulos, Olivier Michielin, Michael Moor , Charlotte Bunne
The Thirty-eighth Annual Conference on Neural Information Processing Systems (NeurIPS 2025)
Multimodal Large Language Models (LLMs) hold promise for biomedical reasoning, but current benchmarks fail to capture the complexity of real-world clinical workflows. Existing evaluations primarily assess unimodal, decontextualized question-answering, overlooking multi-agent decision-making environments such as Molecular Tumor Boards (MTBs). MTBs bring together diverse experts in oncology, where diagnostic and prognostic tasks require integrating heterogeneous data and evolving insights over time. Current benchmarks lack this longitudinal and multimodal complexity. We introduce MTBBench, an agentic benchmark simulating MTB-style decision-making through clinically challenging, multimodal, and longitudinal oncology questions. Ground truth annotations are validated by clinicians via a co-developed app, ensuring clinical relevance. We benchmark multiple open and closed-source LLMs and show that, even at scale, they lack reliability---frequently hallucinating, struggling with reasoning from time-resolved data, and failing to reconcile conflicting evidence or different modalities. To address these limitations, MTBBench goes beyond benchmarking by providing an agentic framework with foundation model-based tools that enhance multi-modal and longitudinal reasoning, leading to task-level performance gains of up to 9.0% and 11.2%, respectively. Overall, MTBBench offers a challenging and realistic testbed for advancing multimodal LLM reasoning, reliability, and tool-use with a focus on MTB environments in precision oncology.
Show BibTeXEeshaan Jain, Johann Wenckstern, Benedikt von Querfurth, Charlotte Bunne
Oral (Top 4%) @ MLGenX Workshop & Poster @ GemBio Workshop, ICLR 2025
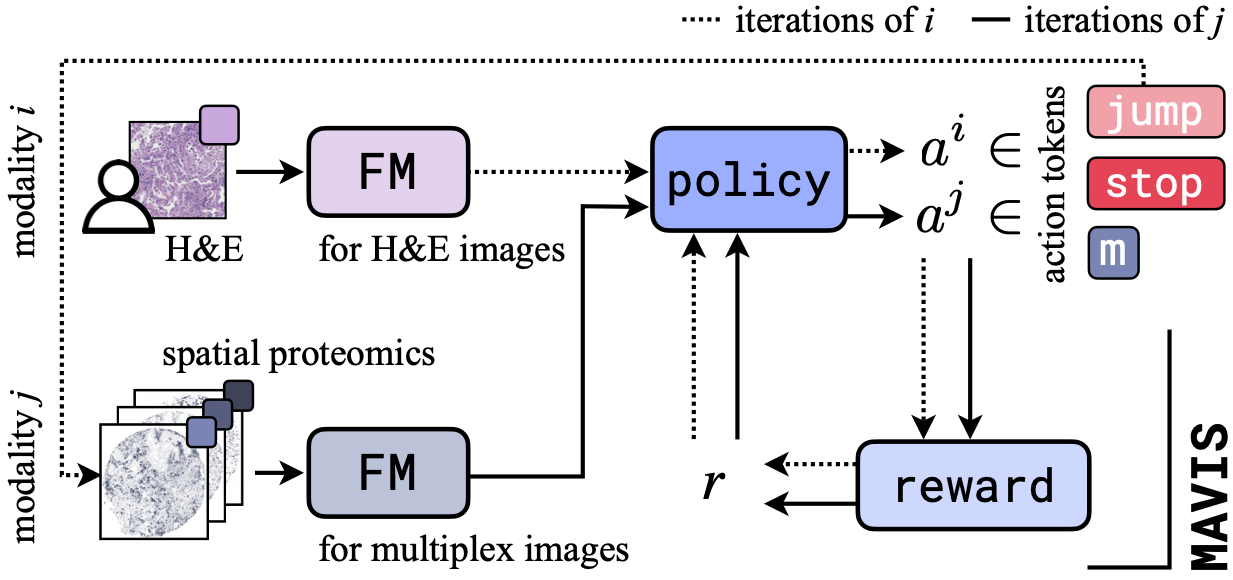
The clinical routine has access to an ever-expanding repertoire of diagnostic tests, ranging from routine imaging to sophisticated molecular profiling technologies. Foundation models have recently emerged as powerful tools for extracting and integrating diagnostic information from these diverse clinical tests, advancing the idea of comprehensive patient digital twins. However, it remains unclear how to select and design tests that ensure foundation models can extract the necessary information for accurate diagnosis. We introduce MAVIS (Multi-modal Active VIew Selection), a reinforcement learning framework that unifies modality selection and feature selection into a single decision process. By leveraging foundation models, MAVIS dynamically determines which diagnostic tests to perform and in what sequence, adapting to individual patient characteristics. Experiments on real-world datasets across multiple clinical tasks demonstrate that MAVIS outperforms conventional approaches in both diagnostic accuracy and uncertainty reduction, while reducing testing costs by over 80%, suggesting a promising direction for optimizing clinical workflows through intelligent test design and selection.
Show BibTeXEeshaan Jain*, Johann Wenckstern*, Kiril Vasilev, Matteo Pariset, Andreas Wicki, Gabriele Gut, Charlotte Bunne
ArXiv Preprint
Spatial proteomics technologies have transformed our understanding of complex tissue architectures by enabling simultaneous analysis of multiple molecular markers and their spatial organization. The high dimensionality of these data, varying marker combinations across experiments and heterogeneous study designs pose unique challenges for computational analysis. Here, we present Virtual Tissues (VirTues), a foundation model framework for biological tissues that operates across the molecular, cellular and tissue scale. VirTues introduces innovations in transformer architecture design, including a novel tokenization scheme that captures both spatial and marker dimensions, and attention mechanisms that scale to high-dimensional multiplex data while maintaining interpretability. Trained on diverse cancer and non-cancer tissue datasets, VirTues demonstrates strong generalization capabilities without task-specific fine-tuning, enabling cross-study analysis and novel marker integration. As a generalist model, VirTues outperforms existing approaches across clinical diagnostics, biological discovery and patient case retrieval tasks, while providing insights into tissue function and disease mechanisms.
Show BibTeXIndradyumna Roy*, Eeshaan Jain*, Soumen Chakrabarti, Abir De
ICLR 2025
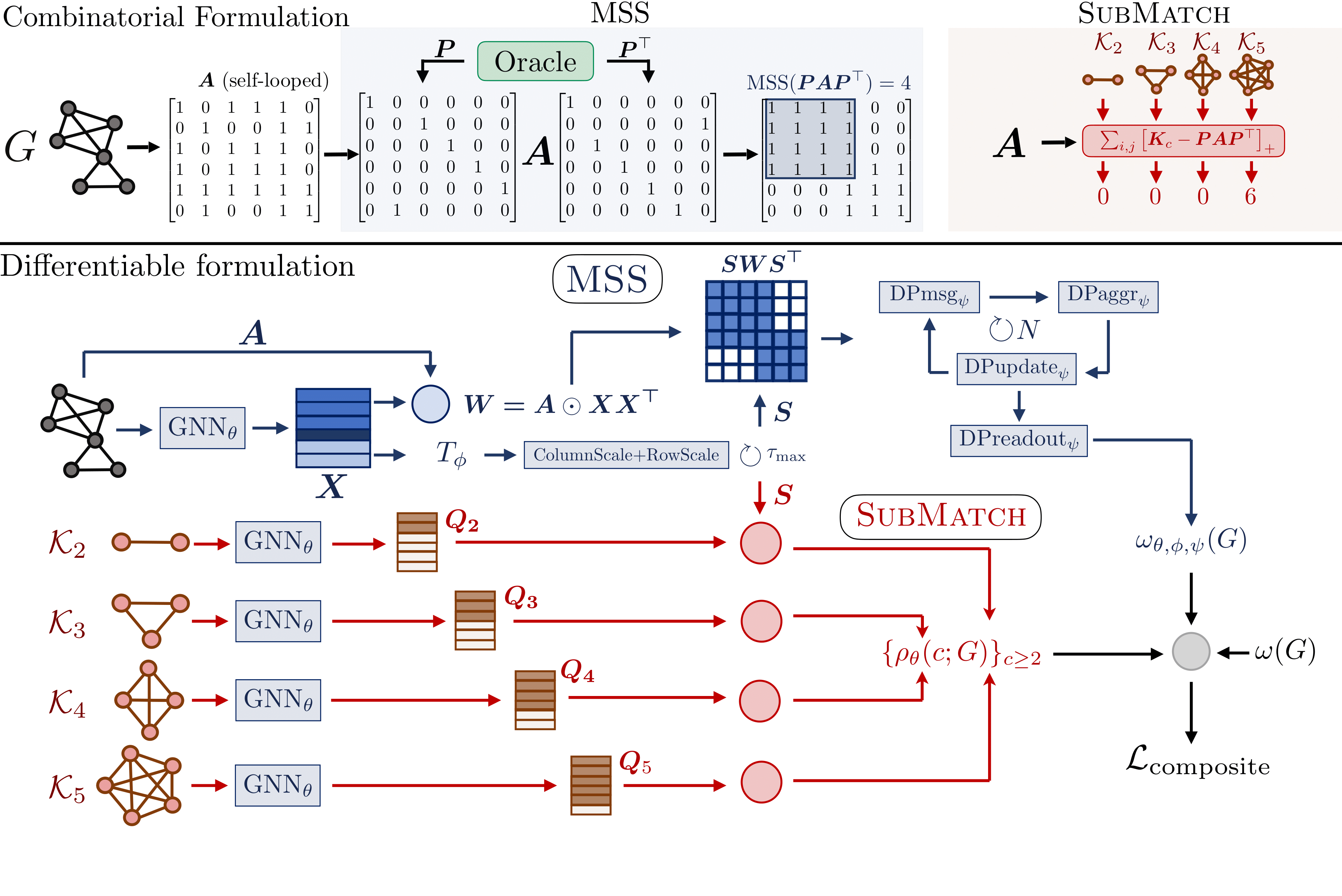
MxNet is a fully differentiable clique number estimator that learns from distant supervision without explicit clique demonstrations. We reformulate MCP as detecting dense submatrices via learned permutations within a nested subgraph matching task.
Show BibTeXEeshaan Jain*, Indradyumna Roy*, Saswat Meher, Soumen Chakrabarti, Abir De
LoG 2024 (Extended Abstract)
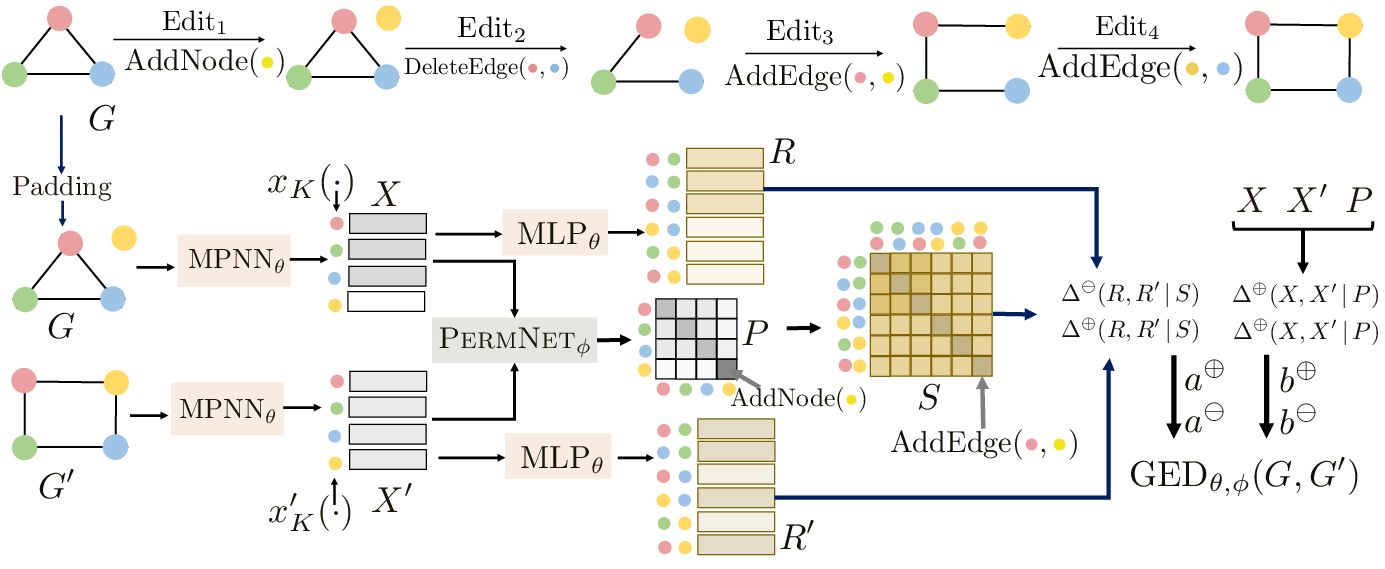
Graph Edit Distance (GED) is a powerful framework for modeling both symmetric and asymmetric relationships between graph pairs under various cost settings. Due to the combinatorial intractability of exact GED computation, recent advancements have focused on neural GED estimators that approximate GED by leveraging data distribution characteristics. However, the datasets commonly used to benchmark such neural models exhibit two critical flaws: (1) significant isomorphism bias and (2) reliance on uniform edit costs for GED ground truths. Our datasets eliminate isomorphism leakage and incorporate a range of edit costs, facilitating more accurate assessment of GED methods.
Show BibTeXEeshaan Jain*, Indradyumna Roy*, Saswat Meher, Soumen Chakrabarti, Abir De
NeurIPS 2024 and LoG 2024 (Extended Abstract)
GraphEdx is the first-of-its-kind neural GED framework that incorporates variable edit costs, capable of modeling both symmetric and asymmetric graph (dis)similarities, allowing for more flexible and accurate GED estimation compared to earlier methods.
Show BibTeXEeshaan Jain, Tushar Nandy, Gaurav Aggarwal, Ashish V. Tendulkar, Rishabh K Iyer, Abir De
NeurIPS 2023
Existing subset selection methods for efficient learning predominantly employ discrete combinatorial and model-specific approaches, which lack generalizability--- for each new model, the algorithm has to be executed from the beginning. We propose `SubSelNet`, a non-adaptive subset selection framework, which tackles these problems.
Show BibTeX